|
|
| POLYPHARMA: A Polypharmacology Database |
| Search Database | Instructions | Citation | Application | Feedback |
|
How to use the Polypharma Database | ||||||||||||||
|
polypharma is a dedicated database for polypharmacological ligands and targets that are crystallized and deposited in PDB, along with their available biological activity data. The database is available at HERE. Below is explained step-by-step procedure to usage of the database The options are available to enter a keyword for either for ligand or protein structure. If the ligand option is chosen | ||||||||||||||
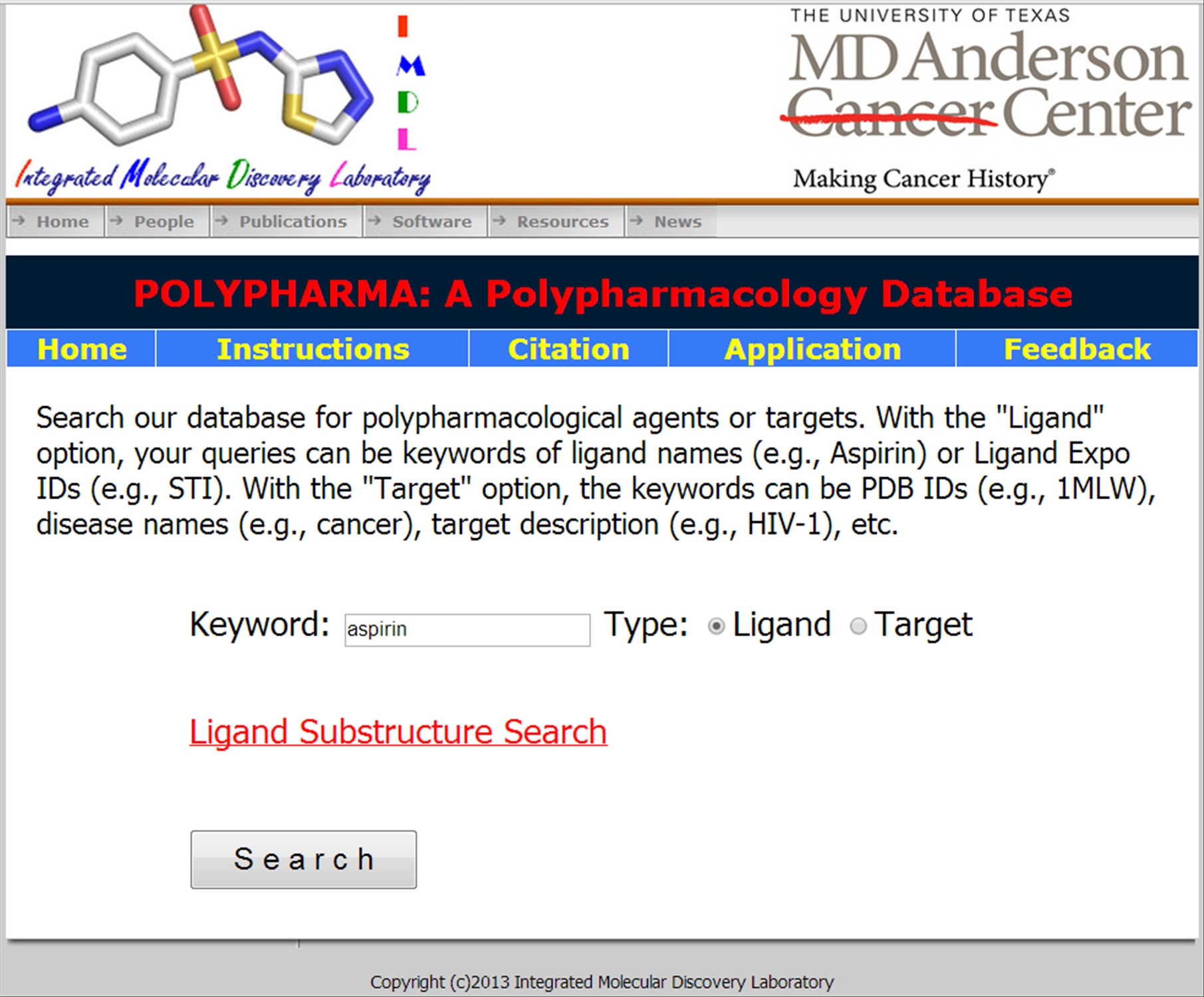 | ||||||||||||||
|
The Ligand option displays all the entries of the ligands (which matched the keyword) along with their identity (ligand_id), 2D image, name, molecular formula, molecular weight, Other names (synonyms). If drugbank entry is available, it also displays Drugbank id, generic and other brand names available. | ||||||||||||||
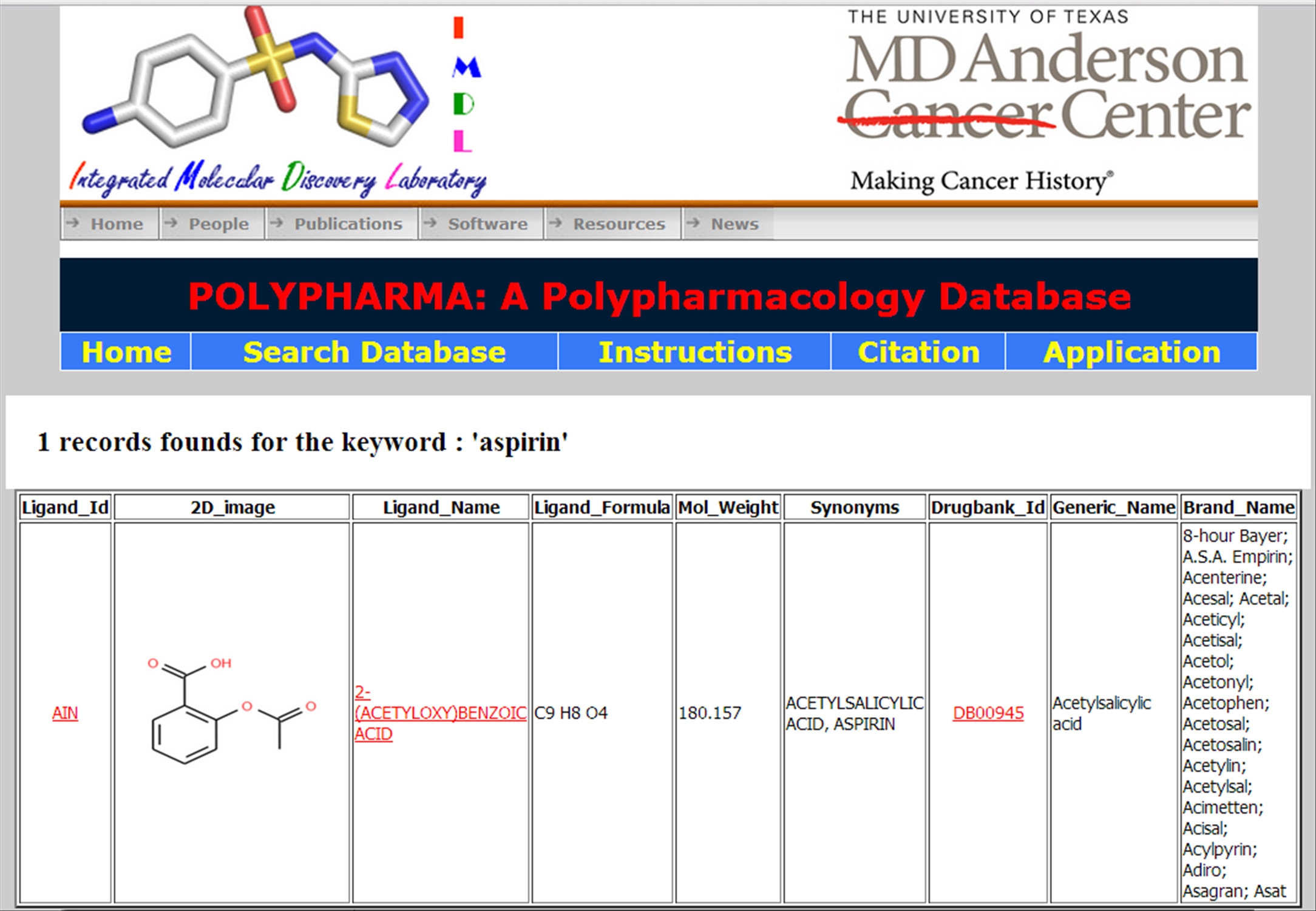
| ||||||||||||||
|
Further clicking on the Lignad_Id displays all the available activity data (Ki, Kd, EC50, IC50, and Ka values) along with its complexed protein information. Ligand_Name is linked to Ligand Expo database for complete details of the ligand. | ||||||||||||||
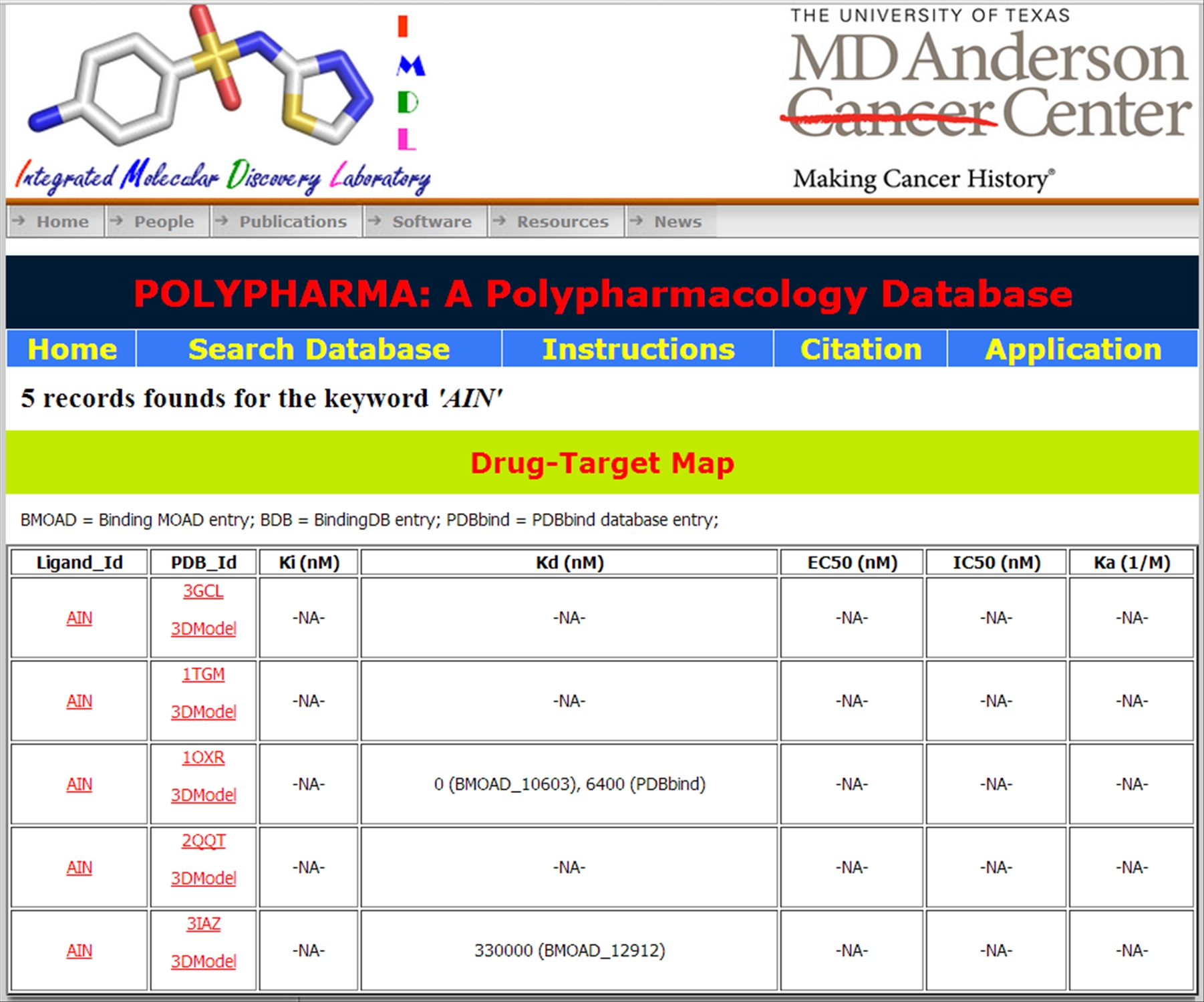
| ||||||||||||||
|
By clicking again on the Ligand_Id we can obtain its detailed inforamtion and properties from the Ligand Expo database. PDB_Id is further hyperlinked to RCSB database for the complete details of the individual protein. The 3D model link displays the 3D structure of the PDB file.
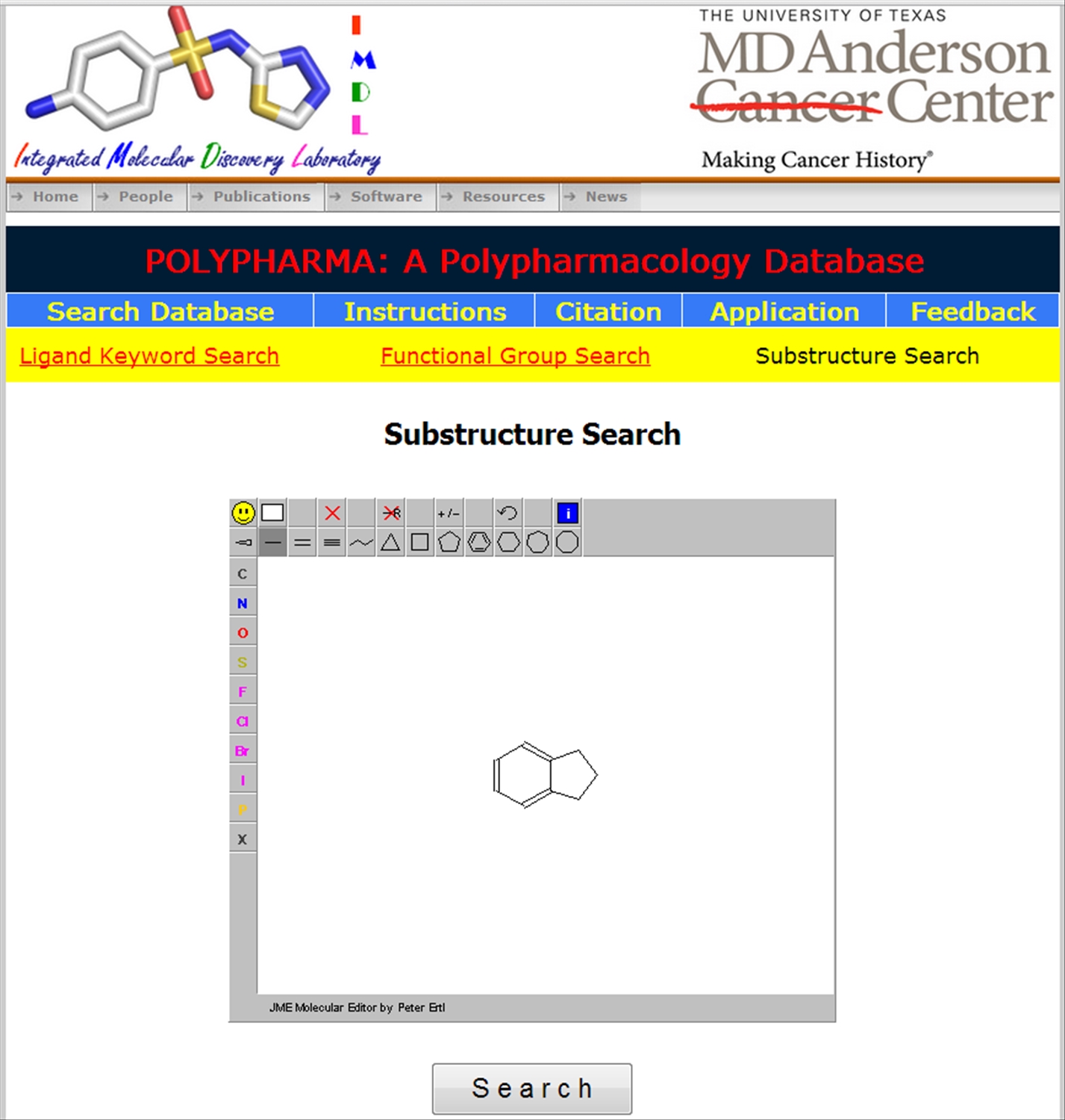
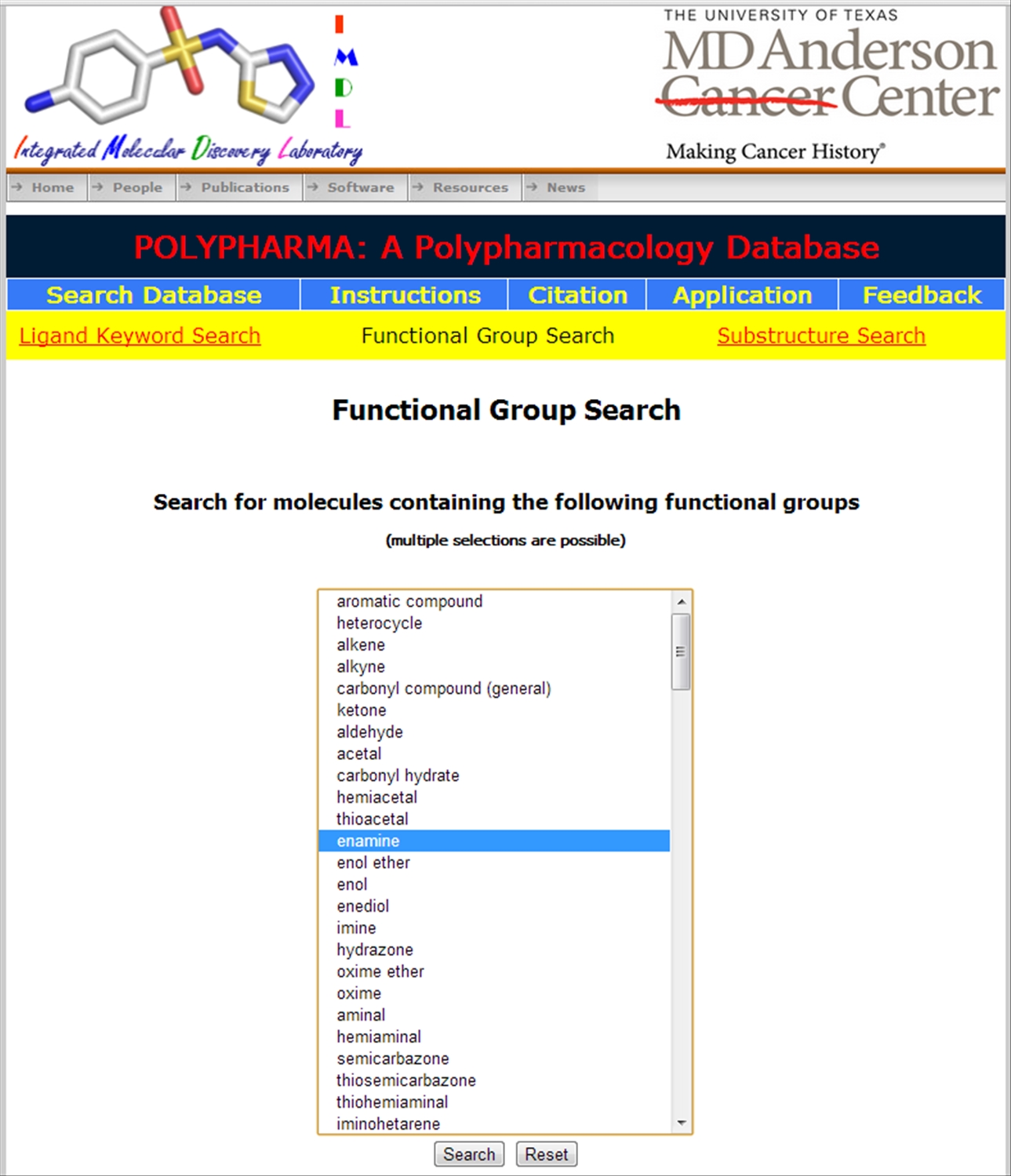
| The polypharmacological ligands can also be searched using (a) either sub-structure search (b) or using functional group
| 
| 
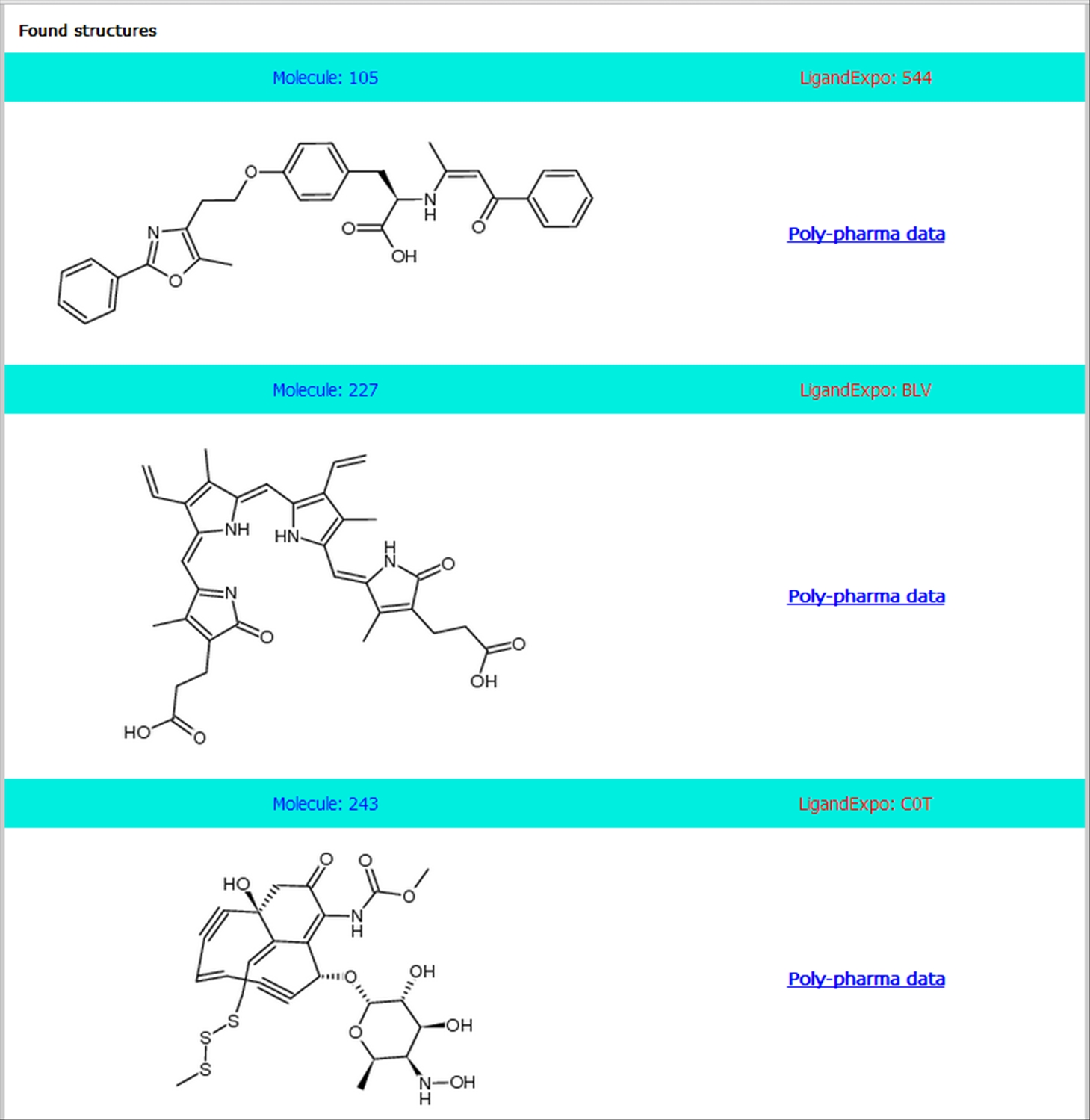
| The results are listed as shown below from either of the searches. Along with the 2D structure of each resulted polypharmacological ligand, links to (a) Ligand's physico-chemical properties (Molecule Id), (b) Original LigandExpo entry page (LigandExpo Id) and (c) available activity data with various targets are provided.
| 
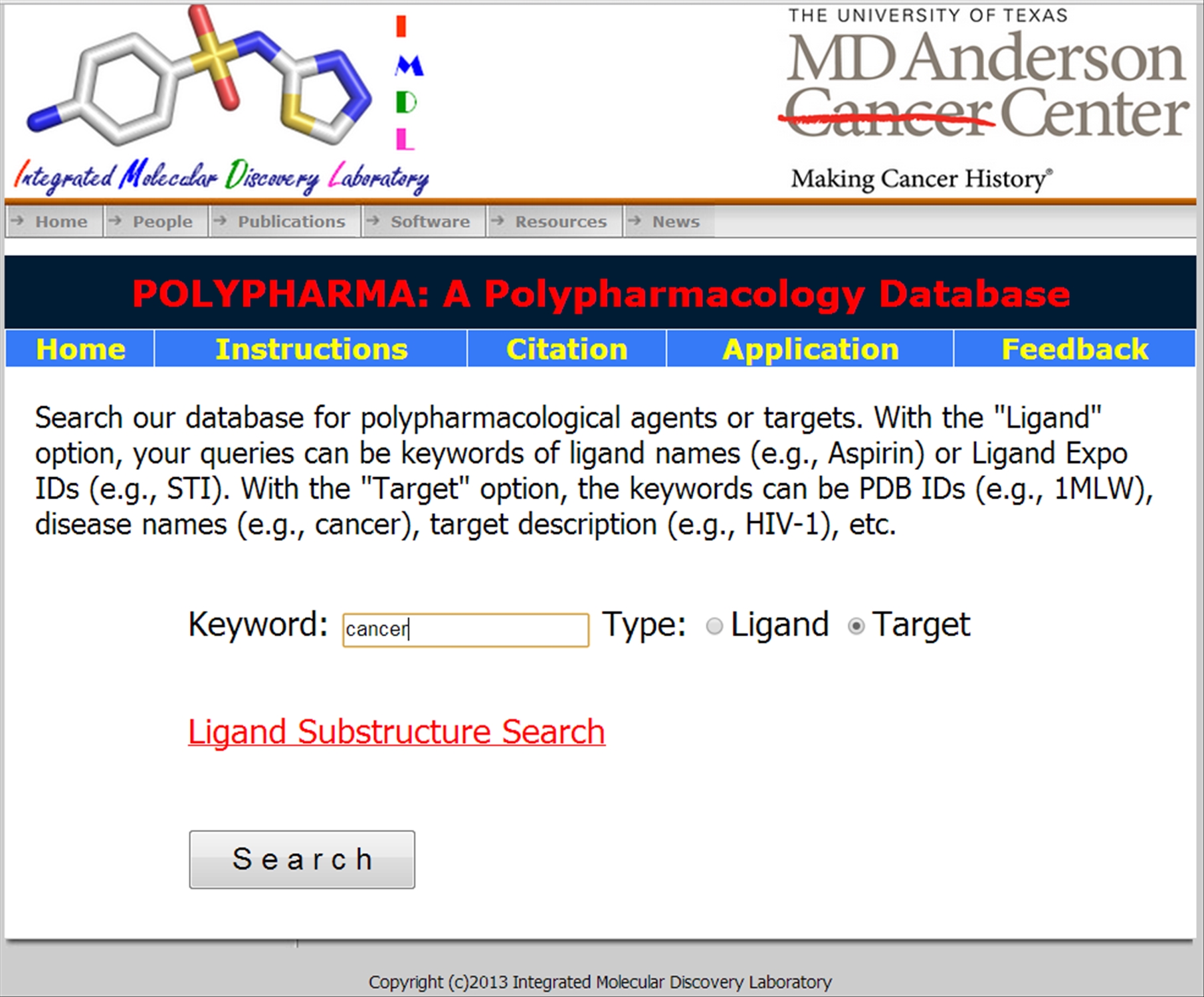
If the Target option is chosen
|
| 
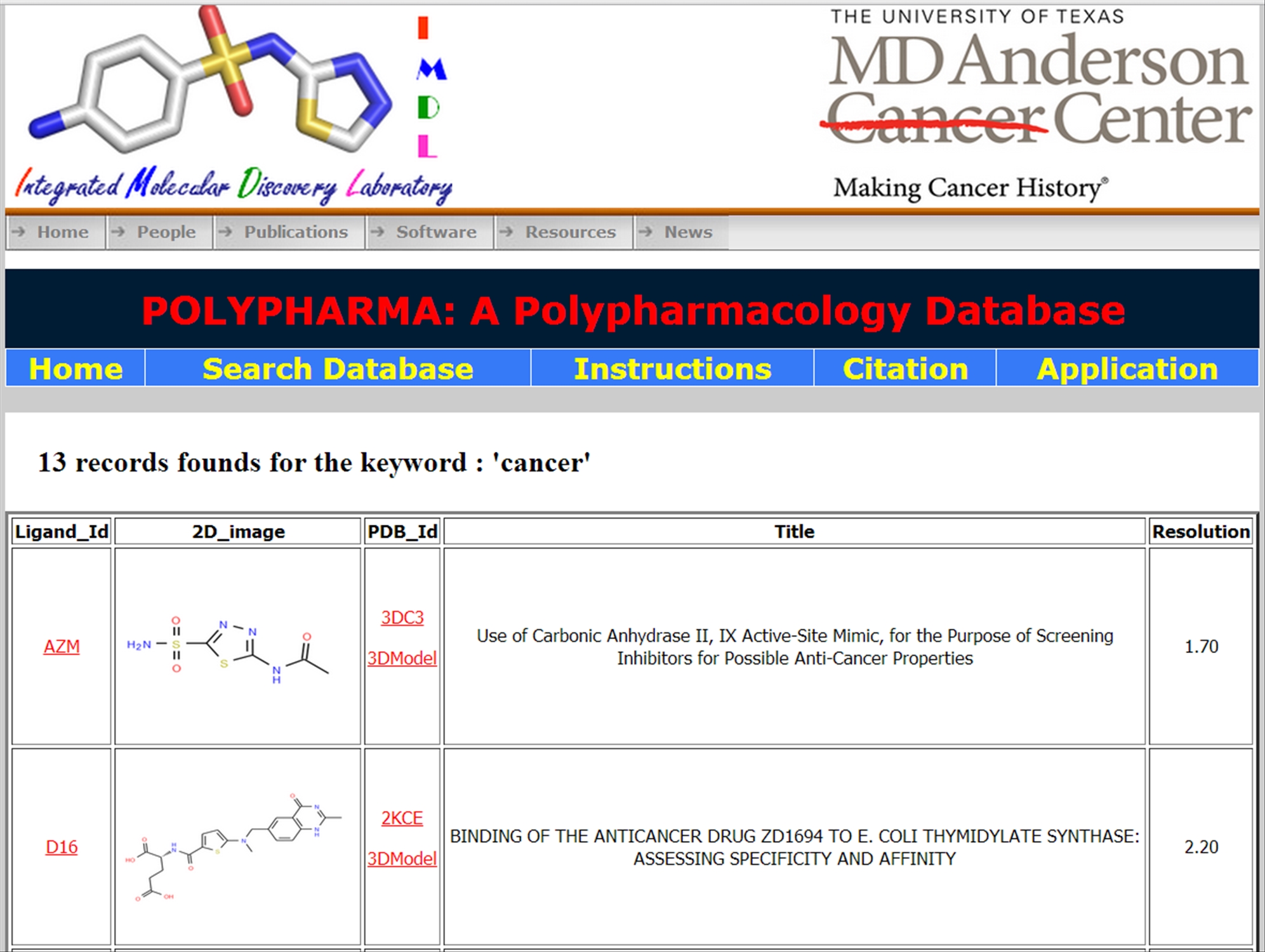
| The Target option displays all the protein structure identities along with their complexed ligand, title and resolution.
| 
| By clicking on the Ligand_Id in protein results screen (shown above), the activity data of the ligand with various bound protein structures can be obtained. PDB_Id is hyperlinked to RCSB database for the complete details of the individual protein.
| 
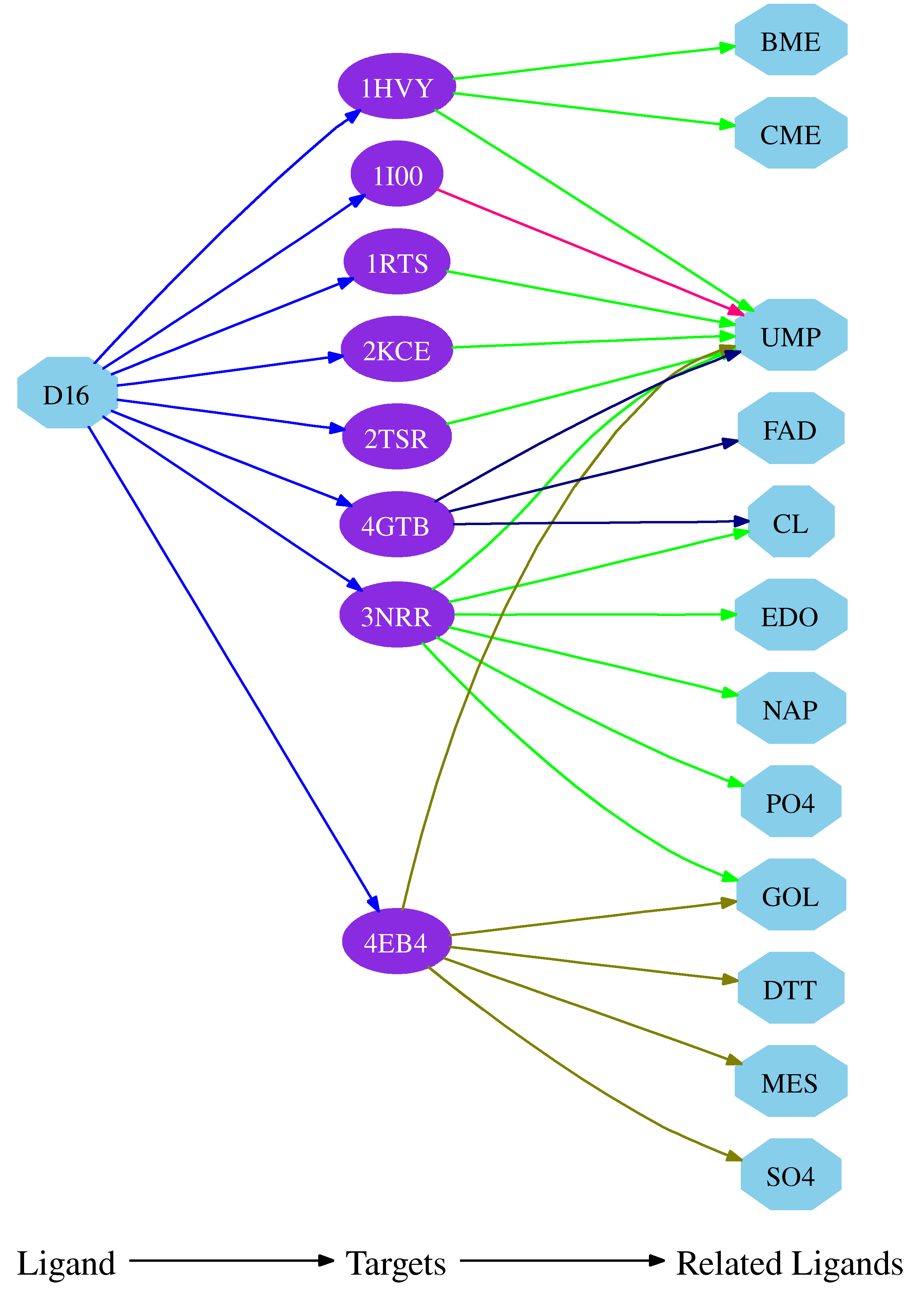
| Drug-Target Map link is provided to graphically obtain the polypharmacological links of the given ligand (DIF in the above example).
| 
|
 |
© 2026 iMDLab |